Grails GORM for Neo4j
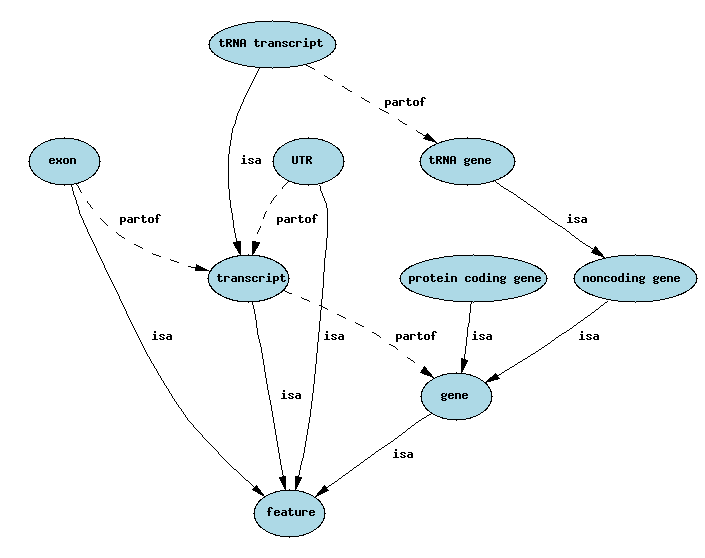
In this post, we are going to make use of the Grails GORM for Neo4j to create a Graph database representing the simplified Sequence Ontology shown in the image below:
NB: Before we get started, I am using the following versions and the IntelliJ IDE:
thoba at xps in ~
$ sdk c
Using:
gradle: 2.14
grails: 3.1.9
groovy: 2.4.7
With that out of the way, let’s get started.
Getting Started
To get started, we need to install the plugin into our Grails application (soneo) by editing the build.gradle file and adding the plugin as a dependency.
dependencies {
compile 'org.grails.plugins:neo4j:5.0.10'
}By default Neo4j will be used in embedded mode, the default directory for the Neo4j datastore is data/neo4j.
Creating our Domains and Controllers
Let’s create our domains and their respective controllers.
By default our classes will be persisted by Hibernate and not Neo4j. To persist our domain classes with Neo4j, we must use the mapWith property:
static mapWith = "neo4j"Domains:
Feature domain:
// Feature domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Feature {
String name
Date created = new Date()
static hasMany = [gene: Gene, exon:Exon, transcript:Transcript, utr:Utr]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Transcript domain:
// Transcript domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Transcript {
String name
Date created = new Date()
static belongsTo = [is_a: Feature, part_of: Gene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Gene domain:
// Gene domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Gene {
String name
Date created = new Date()
static belongsTo = [is_a: Feature]
static hasMany = [coding_gene:Codinggene, noncoding_gene:Noncodinggene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Noncodinggene domain:
// Noncodinggene domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Noncodinggene {
String name
Date created = new Date()
static belongsTo = [is_a: Gene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Codinggene domain:
// Codinggene domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Codinggene {
String name
Date created = new Date()
static belongsTo = [is_a: Gene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Utr domain:
// Utr domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Utr {
String name
Date created = new Date()
static belongsTo = [is_a: Feature, part_of: Transcript]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Exon domain:
// Exon domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class Exon {
String name
Date created = new Date()
static belongsTo = [is_a: Feature, part_of:Transcript]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}TrnaGene domain:
// TrnaGene domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class TrnaGene {
String name
Date created = new Date()
static belongsTo = [is_a: Noncodinggene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}TrnaTranscript domain:
// TrnaTranscript domain
package soneo
import groovy.transform.ToString
@ToString(includes = 'name')
class TrnaTranscript {
String name
Date created = new Date()
static belongsTo = [is_a: Transcript, part_of: TrnaGene]
static constraints = {
name(blank: false)
}
static mapWith = "neo4j"
}Controllers
We will modify our controllers to use dynamic scaffolding, so they will all have the following, replacing {DomainName} with the appropriate domain name:
package soneo
class {DomainName}Controller {
static scaffold = {DomainName}
def index() {}
}With this in place, we can run our app using the grails run-app command, pointing our browser to localhost:8080 and start to populate data.
Viewing our data in Neo4j
In order for us to view our data in Neo4j, we are going to make use of a slightly modified docker image (thoba/neo4j_galaxy_ie:soneo) to cater for user permissions as, by default, docker runs as root.
Also, our application is integrated with Neo4j in embedded mode. This means that one program and only one program can read and write to the database at a time. We will have to stop grails and run the following:
$ cd soneo
$ docker run -d \
-e NEO4J_UID=$(id -u) -e NEO4J_GID=$(id -g) -e NEO4J_AUTH=none \
-v \$(pwd)/data/neo4j:/data \
-p 7474:7474 \
thoba/neo4j_galaxy_ie:soneo
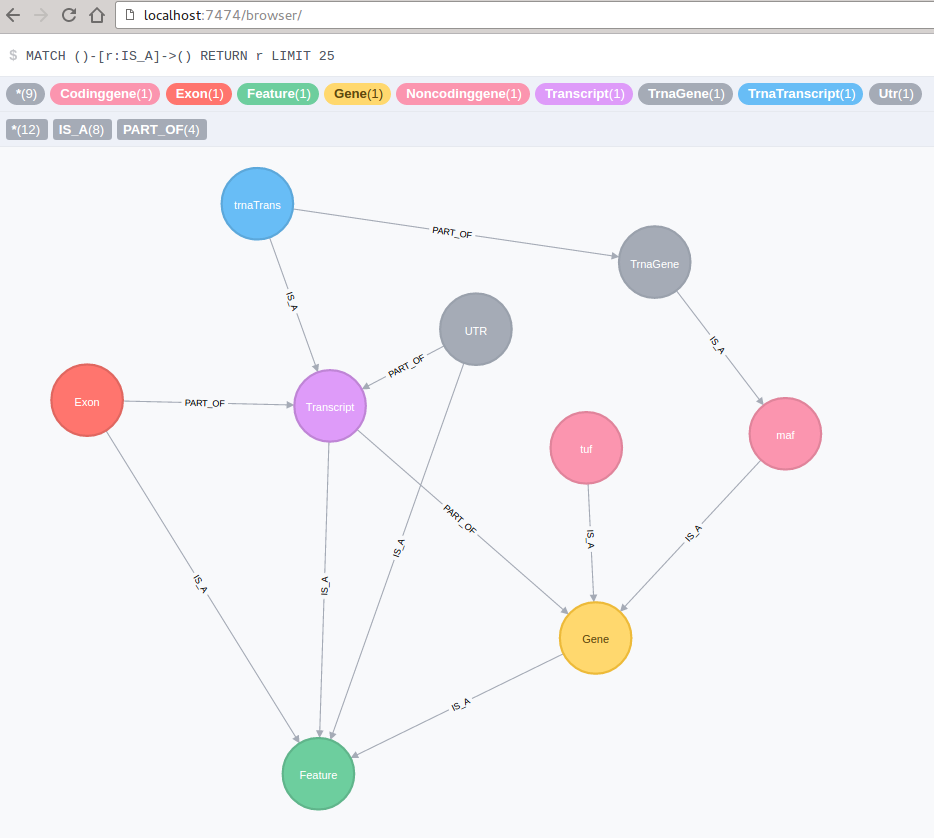
When we see our running container via docker ps, we can point our browser to localhost:7474. Clicking on the IS_A relationship on the side shall present data similar to the graph depicted below:
Code for this post can be found here.

Leave a comment